Sendoel Lab posted the following on X:
“How does aging alter the translational landscape of stem cells and how do these changes impact their regenerative capacity? Here we developed an in vivo single-cell ribosome profiling strategy to monitor translational landscapes of the mouse skin.
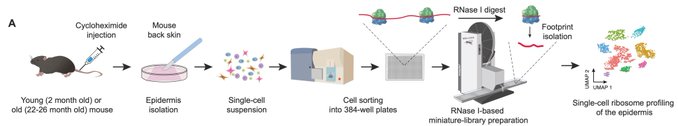
Building on the elegant single-cell protocol by the van Oudenaarden group, we introduced two key modifications: ribosomal elongation-inhibited epidermis isolation and switching to RNase I ribonuclease for generating ribosomal footprints, as commonly used in bulk Ribo-seq.
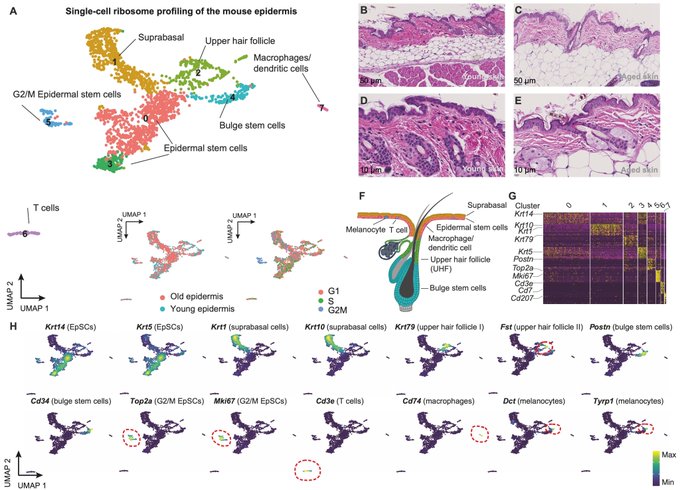
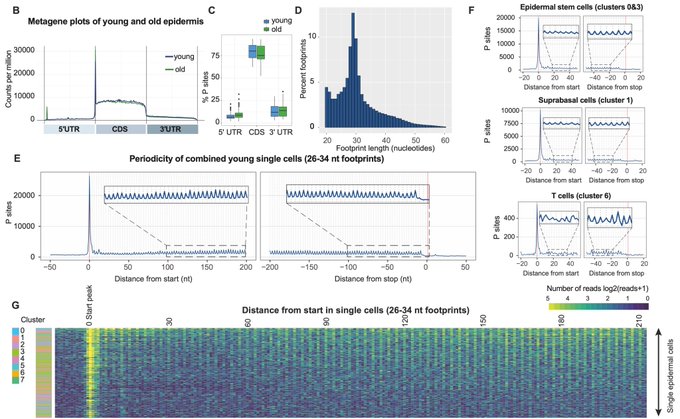
Using our RNase I-based in vivo ribosome profiling strategy, we observed excellent triplet periodicity – a hallmark of high-quality data – across diverse epidermal single cells, including epidermal stem cells, hair follicle cells and T cells.
Combining scRNA-seq and scRibo-seq, we map the in vivo translational landscapes of major epidermal cell types and detail aging-specific shifts in translational efficiencies. We uncover AP-1 reprogramming specifically in aged epidermal stem cells, impacting keratinocyte behavior.
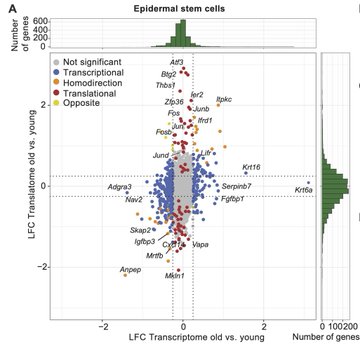
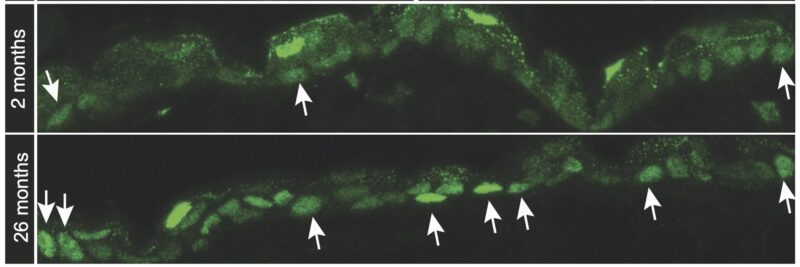
AP-1 levels in aging drive inflammatory responses and inflammaging. AP-1 has also been reported to play a critical role in chromatin opening, thereby altering cellular function and prompting further studies into the posttranscriptional regulation of AP-1 in aged stem cells.
Our study – led by the talented Clara Duré and in collaboration with the Ellis and Levesque labs – suggests a critical role of AP-1 translation in aged stem cells and offers a scalable strategy for tissue-wide interrogation of translational landscapes at single-cell resolution.”
Authors: Clara Duré, Umesh Ghoshdastider, Ramona Weber, Fabiola Valdivia-Francia, Peter F. Renz, Ameya Khandekar, Federica Sella, Katie Hyams, David Taborsky, Merve Yigit, Mark Ormiston, Homare Yamahachi, Mitchell Levesque, Stephanie Ellis, Ataman Sendoel

