Andy Zeng, MD/PhD Candidate at the University of Toronto, shared on X:
“I’m excited to share an R package that we’ve created for mapping scRNA-seq data across normal and malignant hematopoiesis! We’ve used this to identify hematopoietic stem cells from healthy tissues and map leukemia cells from hundreds of patients.
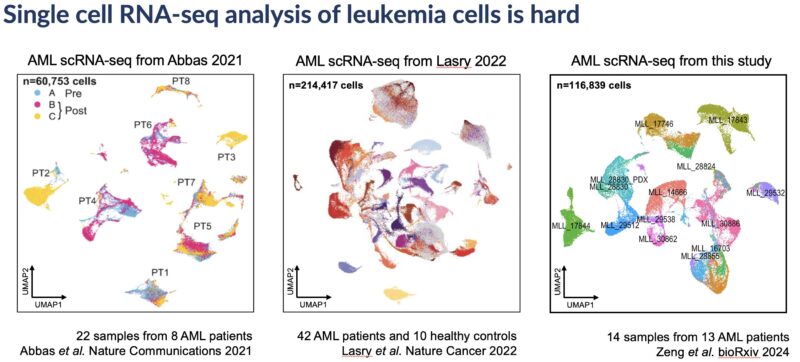
If you’ve performed scRNA-seq on leukemia samples, you’ll be know that analysis can be a chaotic process. Inter-patient heterogeneity dominates clustering even after batch correction, and cell type annotation of individual leukemia cells is also really difficult.
So, we decided to create a scRNA-seq reference atlas with even proportions of primitive stem and progenitor cells and differentiated immune cells. Importantly, we annotated the cells to be concordant with FACS-purified HSPC fractions, as well as highly purified stem cells.

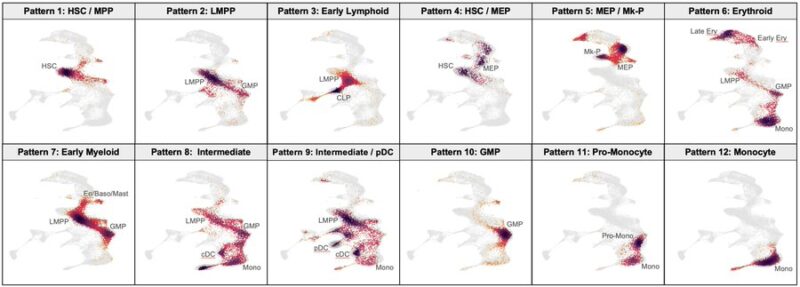
This map can be used to visualize how hematopoiesis becomes distorted in leukemia. We mapped scRNA-seq data from hundreds of AML patients and identified recurrent patterns of AML differentiation. These were linked to underlying genetic mutations.
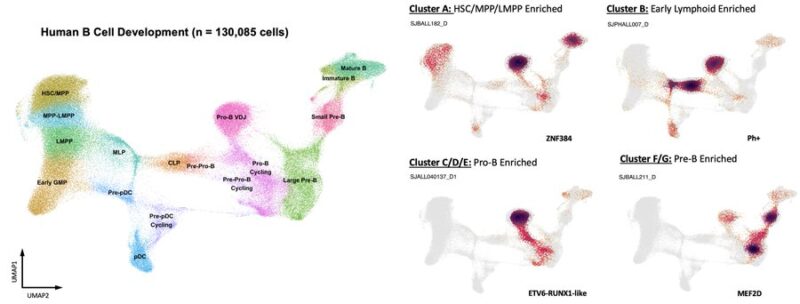
We’ve also developed a focused map of human B-cell development spanning fetal and post-natal tissues, used to map 89 B-ALL patient samples. While most cells were Pro-B, there was involvement of multipotent cell states in a subset of B-ALL patients.
Our tutorials teach you to map scRNA-seq data from normal and malignant blood samples within minutes.
Alternatives:
I don’t like Azimuth due to low HSPC resolution, but here are some other great atlases:
An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors.”